For classic binary classification problems, learning algorithms mostly need extended features for both positive class and negative class. Unfortunately a significant number of applications involve only positive cases. In regulatory Genomics, researchers found the positive cases through wet lab experiments and large scale post-GWAS analysis leading to extremely high confidence that a SNP (Single Nucleotide Polymorphism) is the functional genome with label as positive. However, for most of time, it is difficult to statistically proof specific genetic factor is NOT a functional one.
In order to train binary classification models, traditional learning algorithms accept the data unexplored as negative cases and train on them. Undoubtfuly, training set generated in this way is not reliable and potentially causes lots of bias for both training and classification. Thus, we have to get along with unlabeled data and try our best to classify future cases.
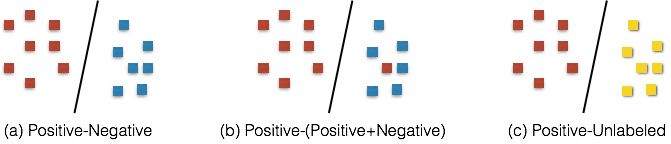
Generally, the rest data (accessible data out of already-known positive cases) are merged to training set as “unlabeled dataset”. Another preprocess frequently used is to apply an implicit criterion to generate “negative” data which actually contains both positive and negative cases in it. This filtering criterion is based on domain-specific knowledge that, there will be less positive cases in unlabeled dataset after filtering. This operation makes the unlabeled dataset more close to “pure” negative dataset.
Reference
[1] Elkan, C., & Noto, K. (2008). Learning classifiers from only positive and unlabeled data (pp. 213–9). Presented at the the 14th ACM SIGKDD international conference, New York, New York, USA: ACM Press.
